Describing Data 👨💻
MATH 4720/MSSC 5720 Introduction to Statistics
Department of Mathematical and Statistical Sciences
Marquette University
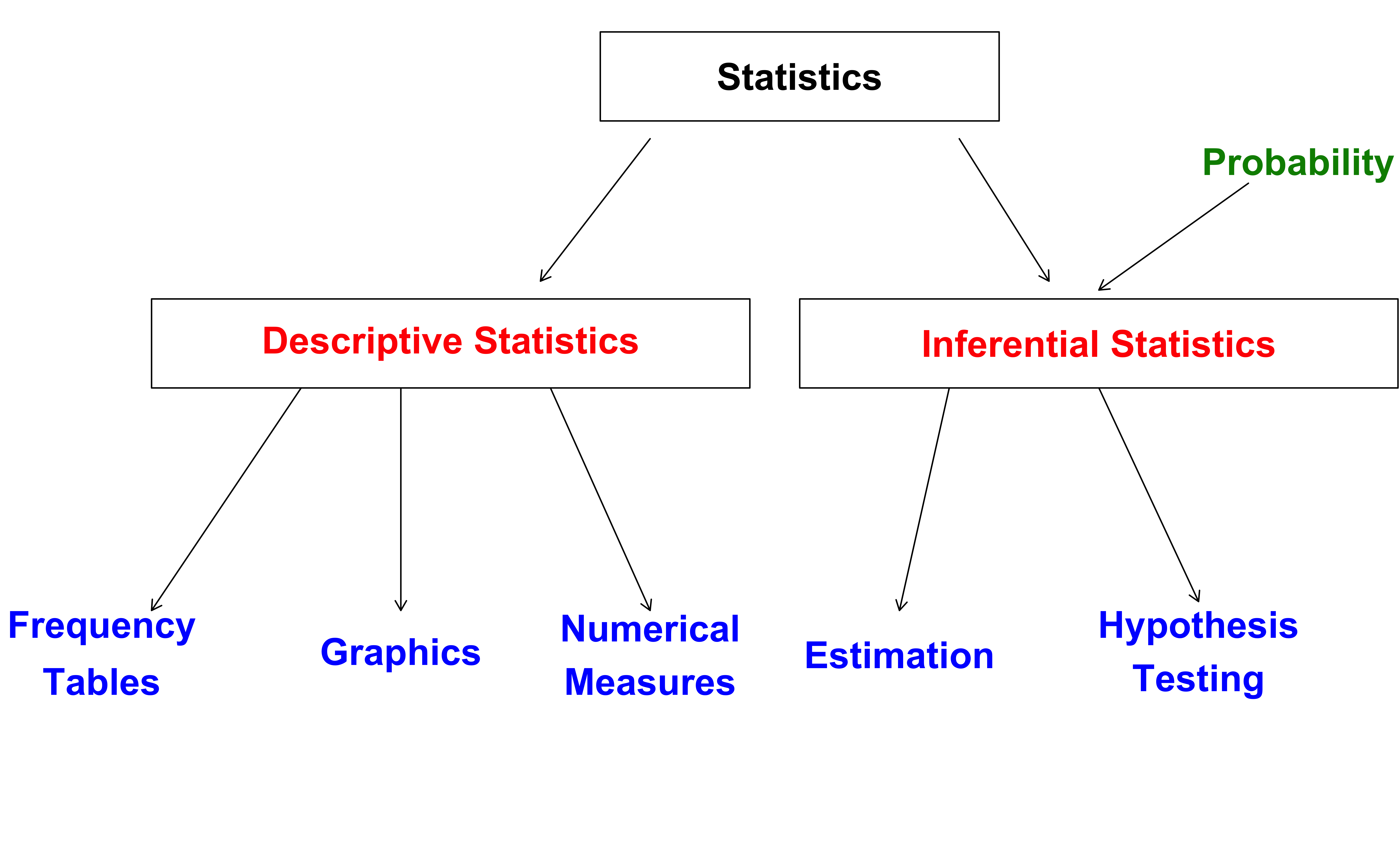
Descriptive Statistics
Data Summary in Tables, Graphics and Numerical Values
Descriptive Statistics (Data Summary)
- Before doing inferential statistics, let’s first learn to understand our data by describing or summarizing it using a table, graph, or some important measures, so that appropriate methods can be performed for better inference results.
Frequency Table for Categorical Variable
A frequency table (frequency distribution) lists variable values individually for categorical data along with their corresponding number of times occurred in the data (frequencies or counts).
Frequency table for categorical data with \(n\) data values:
| Category name | Frequency | Relative Frequency |
|---|---|---|
| \(C_1\) | \(f_1\) | \(f_1/n\) |
| \(C_2\) | \(f_2\) | \(f_2/n\) |
| … | … | … |
| \(C_k\) | \(f_k\) | \(f_k/n\) |
Frequency Table for Categorical Variable
A frequency table (frequency distribution) lists variable values individually for categorical data along with their corresponding number of times occurred in the data (frequencies or counts).
Example: A categorical variable color that has three categories
| Category name | Frequency | Relative Frequency |
|---|---|---|
| Red 🔴 | 8 | 8/50 = 0.16 |
| Blue 🔵 | 26 | 26/50 = 0.52 |
| Black ⚫ | 16 | 16/50 = 0.32 |
R Packages 📦
Packages wrap up reusable R functions, the documentation that describes how to use them, and data sets all together.
As of August 2025, there are about 22510 R packages available on CRAN (the Comprehensive R Archive Network)!
Categorical Frequency Table palmerpenguins package ![]()
str(penguins)tibble [344 × 8] (S3: tbl_df/tbl/data.frame)
$ species : Factor w/ 3 levels "Adelie","Chinstrap",..: 1 1 1 1 1 1 1 1 1 1 ...
$ island : Factor w/ 3 levels "Biscoe","Dream",..: 3 3 3 3 3 3 3 3 3 3 ...
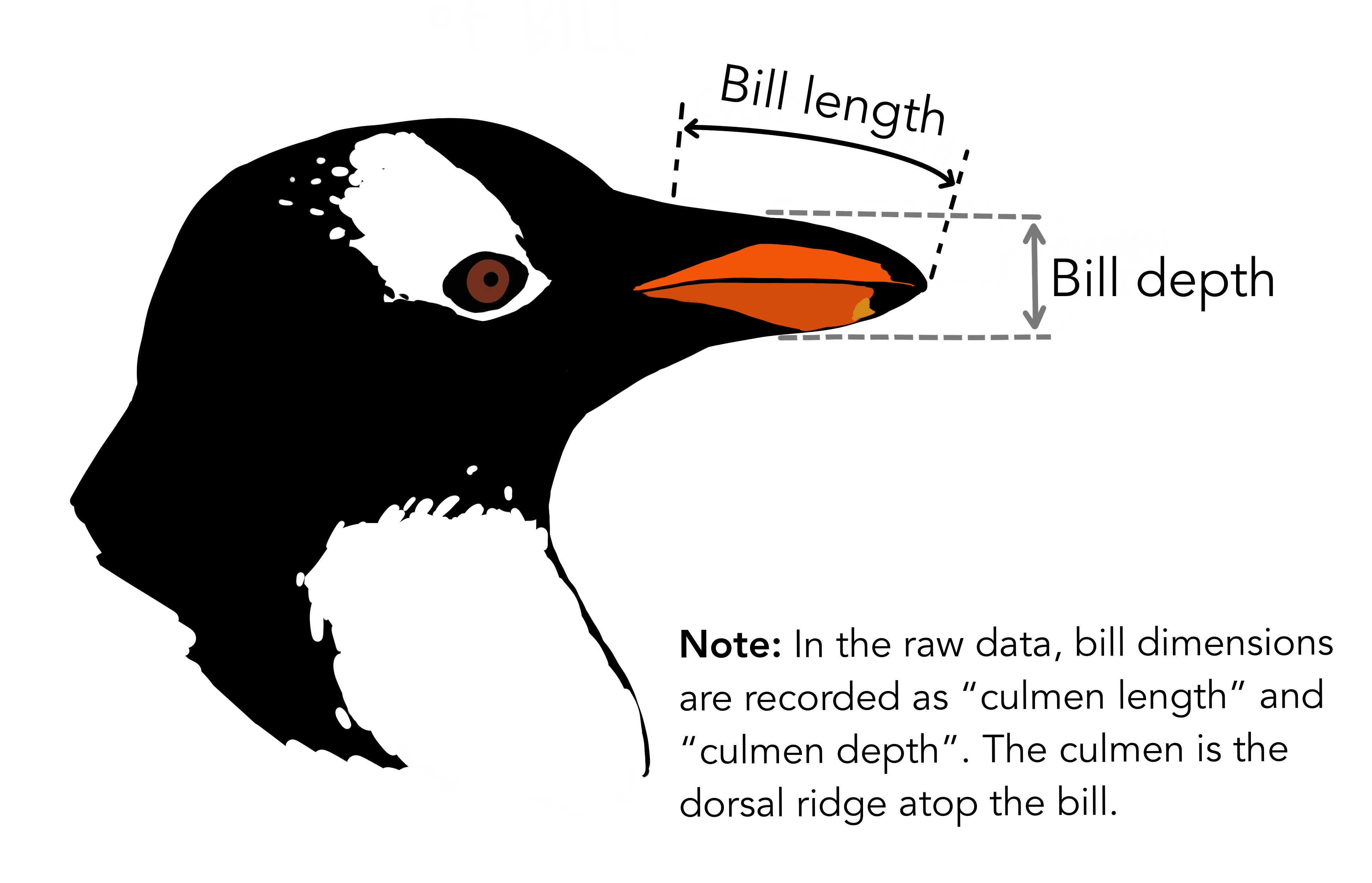
$ bill_length_mm : num [1:344] 39.1 39.5 40.3 NA 36.7 39.3 38.9 39.2 34.1 42 ...
$ bill_depth_mm : num [1:344] 18.7 17.4 18 NA 19.3 20.6 17.8 19.6 18.1 20.2 ...
$ flipper_length_mm: int [1:344] 181 186 195 NA 193 190 181 195 193 190 ...
$ body_mass_g : int [1:344] 3750 3800 3250 NA 3450 3650 3625 4675 3475 4250 ...
$ sex : Factor w/ 2 levels "female","male": 2 1 1 NA 1 2 1 2 NA NA ...
$ year : int [1:344] 2007 2007 2007 2007 2007 2007 2007 2007 2007 2007 ...x <- penguins[, "species"]Categorical Frequency Table: species
## frequency table
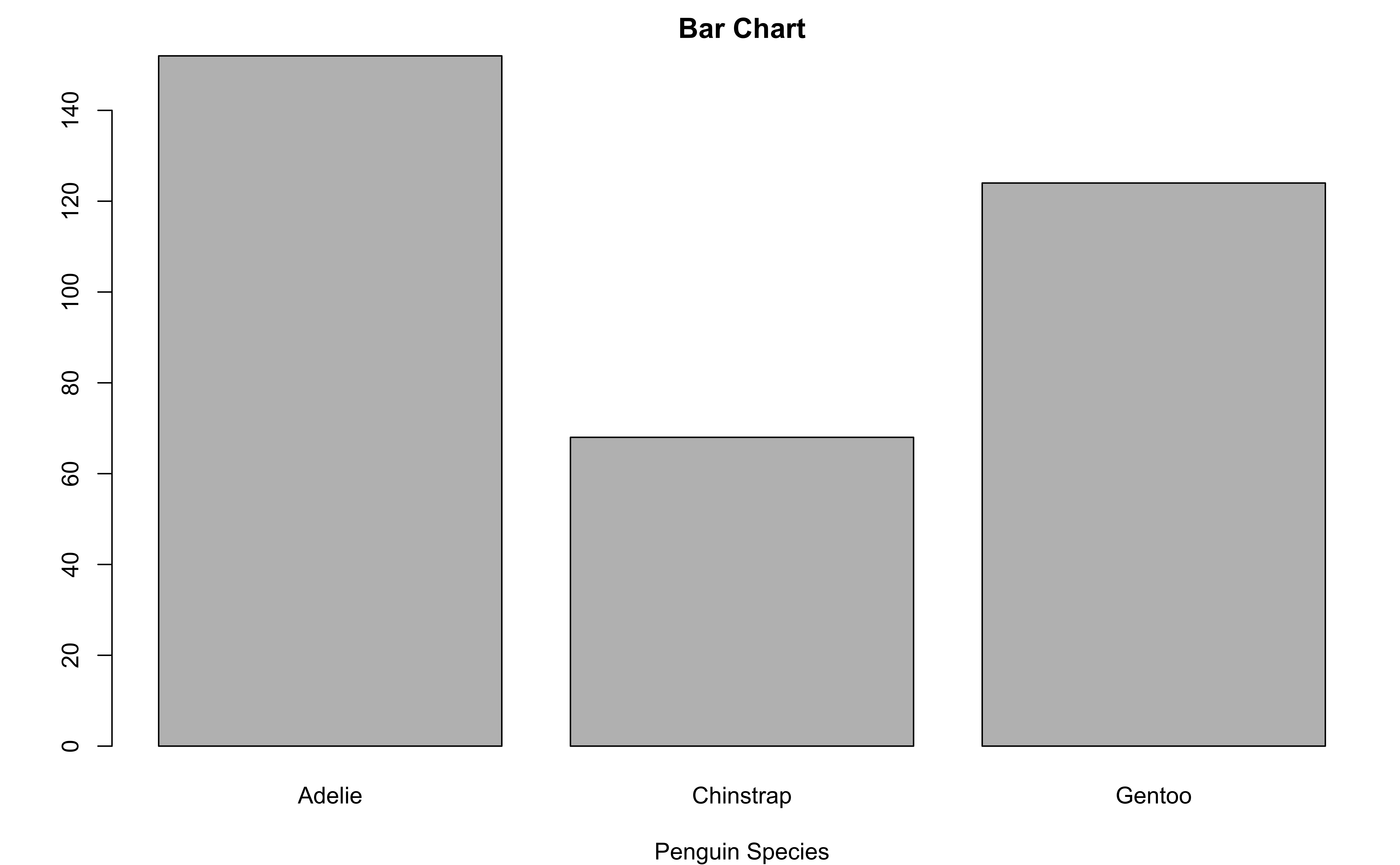
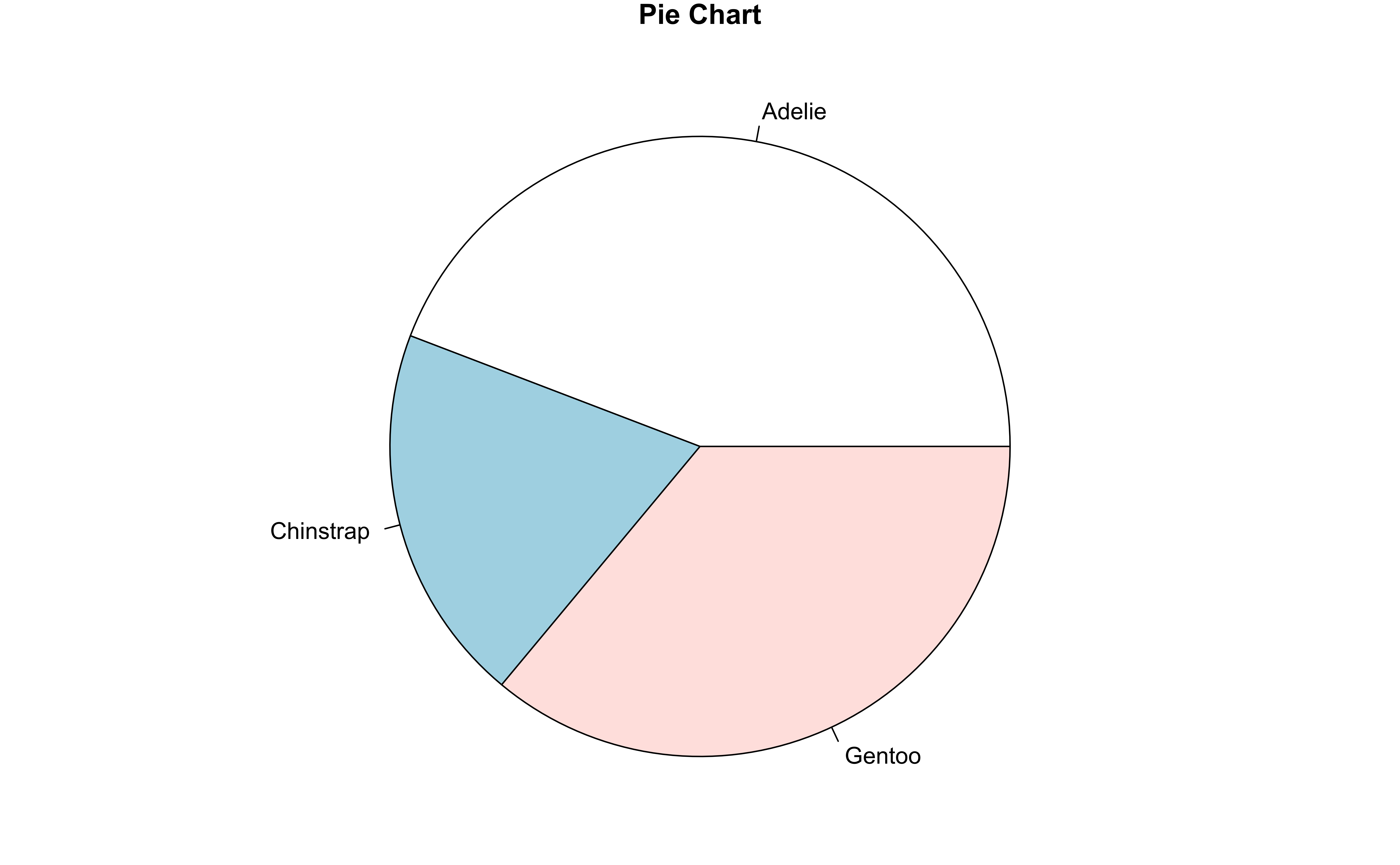
table(x)species
Adelie Chinstrap Gentoo
152 68 124 Visualizing a Frequency Table: Bar Chart
Pie Chart
Frequency Distribution for Numerical Variables
Divide the data into \(k\) non-overlapping groups of intervals (classes).
Convert the data into \(k\) categories with an associated class interval.
Count the number of measurements falling in a given class interval (class frequency).
| Class | Class Interval | Frequency | Relative Frequency |
|---|---|---|---|
| \(1\) | \([a_1, a_2]\) | \(f_1\) | \(f_1/n\) |
| \(2\) | \((a_2, a_3]\) | \(f_2\) | \(f_2/n\) |
| … | … | … | … |
| \(k\) | \((a_k, a_{k+1}]\) | \(f_k\) | \(f_k/n\) |
- \((a_2 - a_1) = (a_3 - a_2) = \cdots = (a_{k+1} - a_k)\). All class widths are the same!
Frequency Distribution for Numerical Variables
Divide the data into \(k\) non-overlapping groups of intervals (classes).
Convert the data into \(k\) categories with an associated class interval.
Count the number of measurements falling in a given class interval (class frequency).
| Class | Class Interval | Frequency | Relative Frequency |
|---|---|---|---|
| \(1\) | \([80, 100]\) | \(2\) | \(2/50\) |
| \(2\) | \((100, 120]\) | \(4\) | \(4/50\) |
| … | … | … | … |
| \(8\) | \((220, 240]\) | \(3\) | \(3/50\) |
Can our grade conversion be used for creating a frequency distribution?
| Grade | Percentage |
|---|---|
| A | [94, 100] |
| A- | [90, 94) |
| B+ | [87, 90) |
| B | [83, 87) |
| B- | [80, 83) |
| C+ | [77, 80) |
| C | [73, 77) |
| C- | [70, 73) |
| D+ | [65, 70) |
| D | [60, 65) |
| F | [0, 60) |
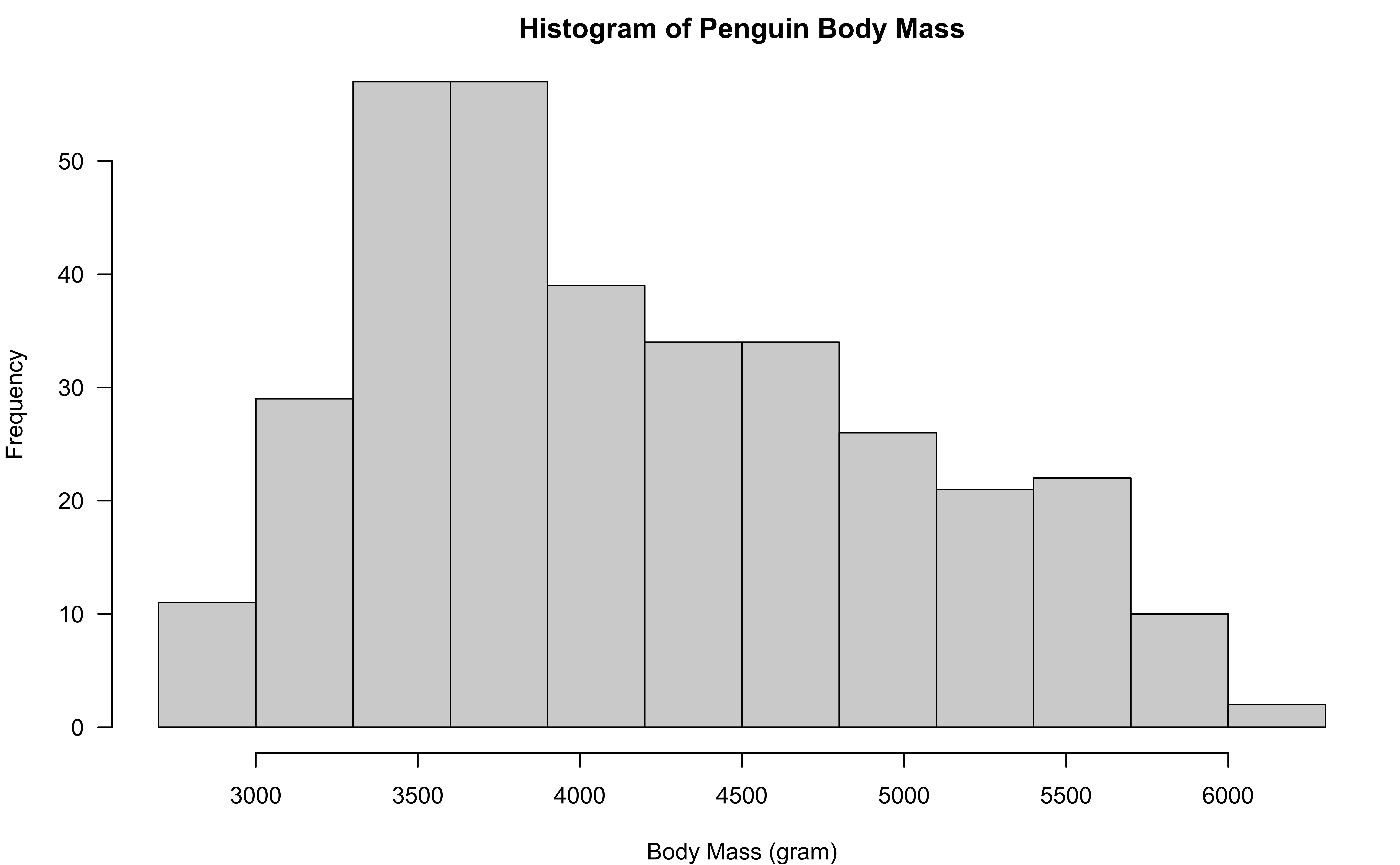
Body Mass (Grams) in Data penguins
body_mass <- penguins$body_mass_g
head(body_mass, 20) [1] 3750 3800 3250 NA 3450 3650 3625 4675 3475 4250 3300 3700 3200 3800 4400
[16] 3700 3450 4500 3325 4200body_mass <- body_mass[complete.cases(body_mass)]Frequency Distribution of Body Mass
Class Class_Intvl Freq Rel_Freq
1 2700g - 3000g 11 0.03
2 3000g - 3300g 29 0.08
3 3300g - 3600g 57 0.17
4 3600g - 3900g 57 0.17
5 3900g - 4200g 39 0.11
6 4200g - 4500g 34 0.10
7 4500g - 4800g 34 0.10
8 4800g - 5100g 26 0.08
9 5100g - 5400g 21 0.06
10 5400g - 5700g 22 0.06
11 5700g - 6000g 10 0.03
12 6000g - 6300g 2 0.01range(body_mass)[1] 2700 6300- All class widths are the same!
- Number of classes should not be too big or too small.
- The lower limit of the 1st class should not be greater than the minimum value of the data.
- The upper limit of the last class should not be smaller than the maximum value of the data.
Wonder how we choose the number of classes or the class width?
R decide the number for us when we visualize the frequency distribution by a histogram.
Visualizing Frequency Distribution by a Histogram
Use default breaks (no need to specify)
hist(x = body_mass,
xlab = "Body Mass (gram)",
main = "Histogram (Defualt)")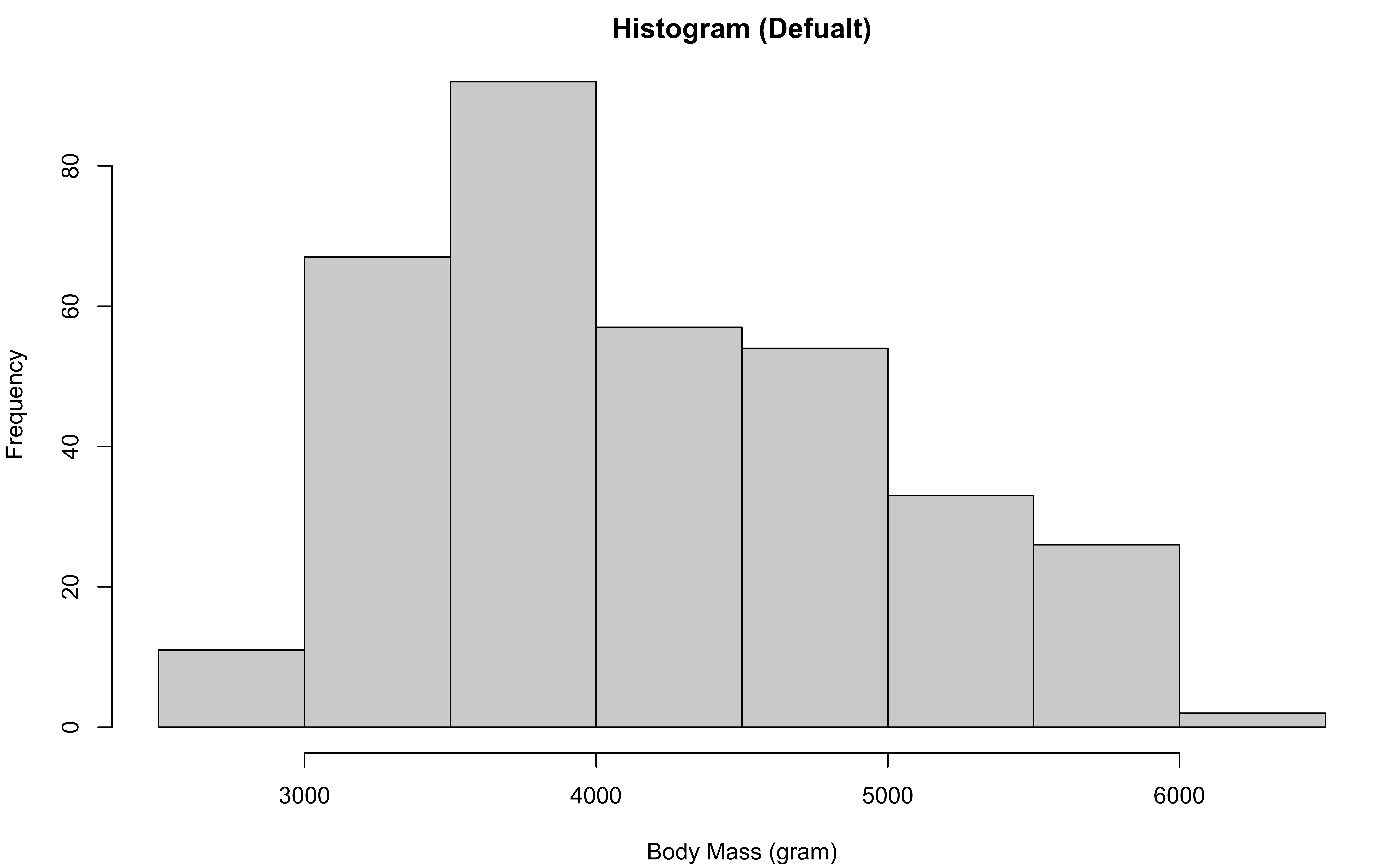
Use customized breaks
class_boundary [1] 2700 3000 3300 3600 3900 4200 4500 4800 5100 5400 5700 6000 6300hist(x = body_mass,
breaks = class_boundary, #<<
xlab = "Body Mass (gram)",
main = "Histogram (Ours)")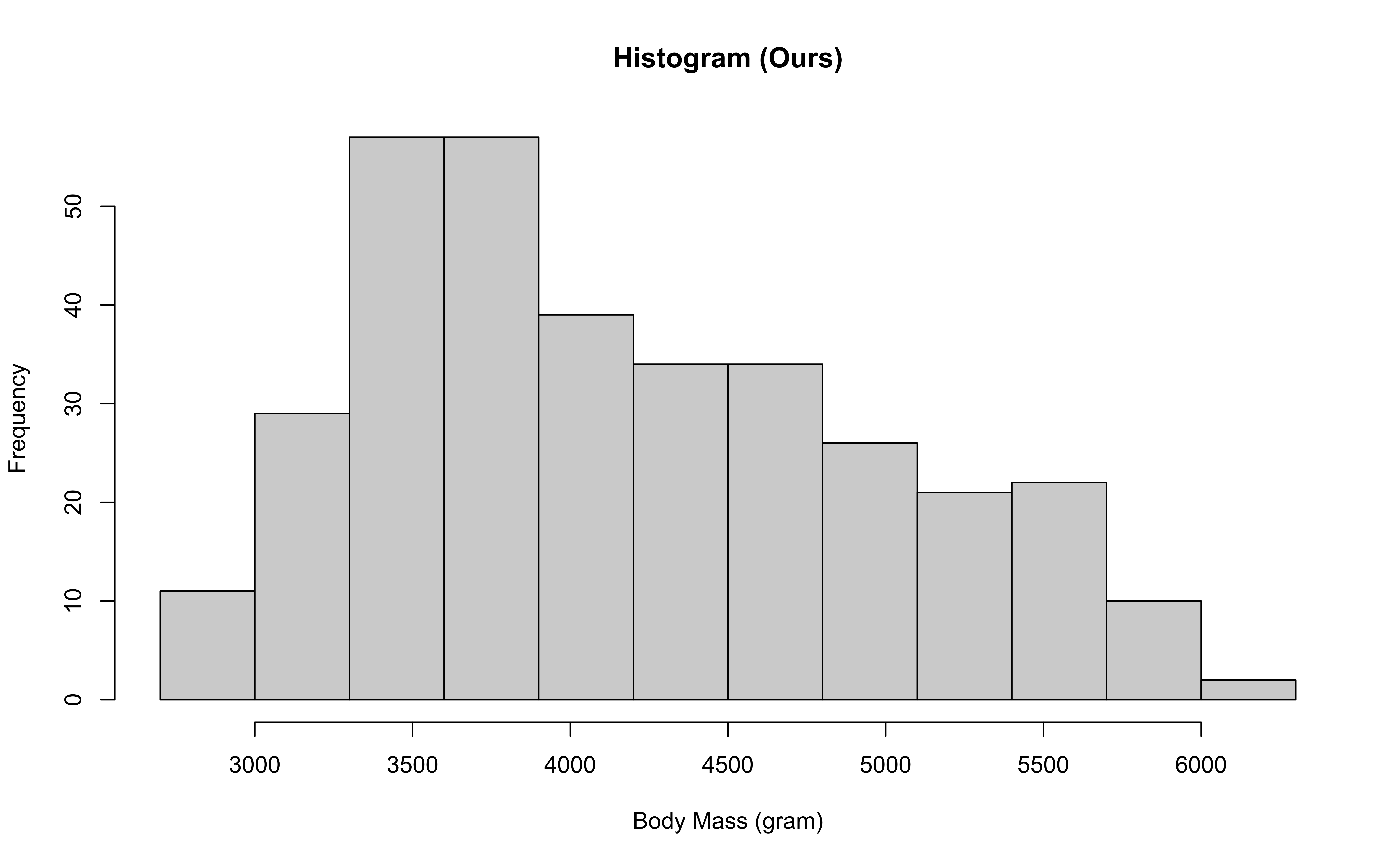
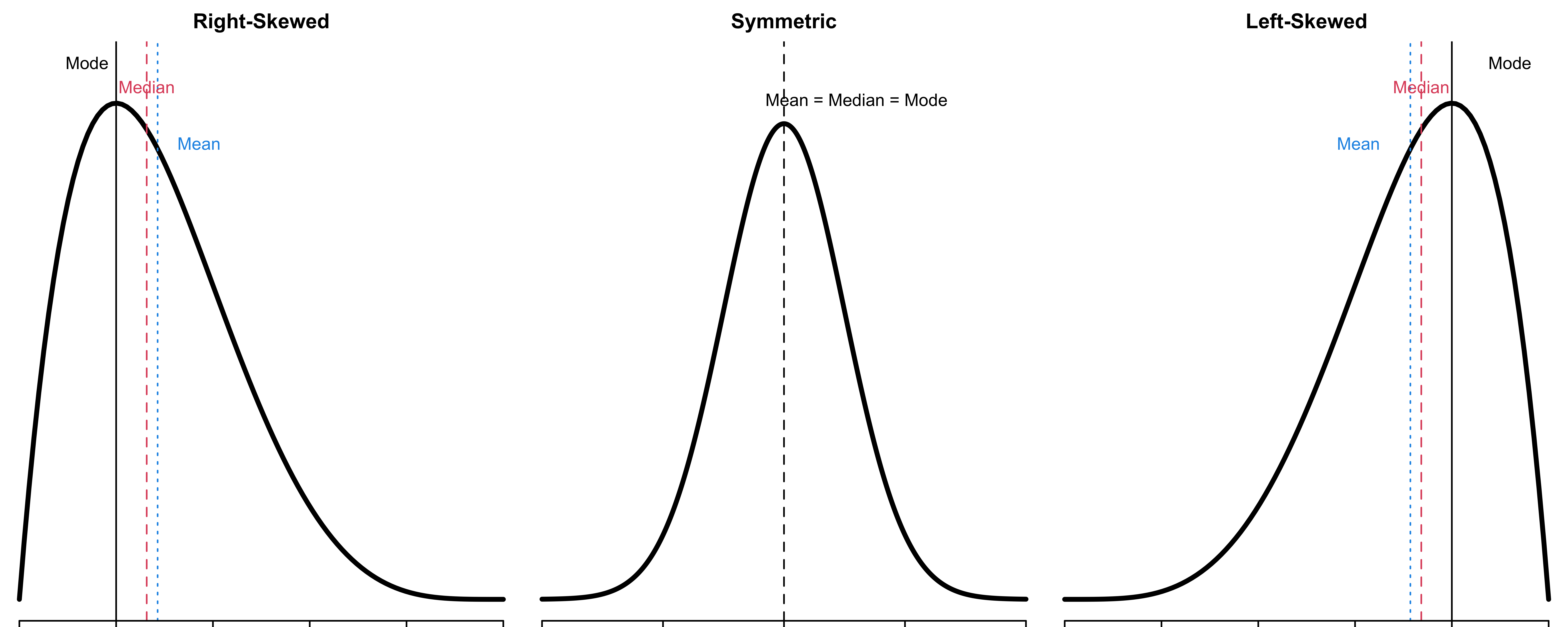
Skewness
Key characteristics of distributions includes shape, center and spread.
Skewness provides a way to summarize the shape of a distribution.
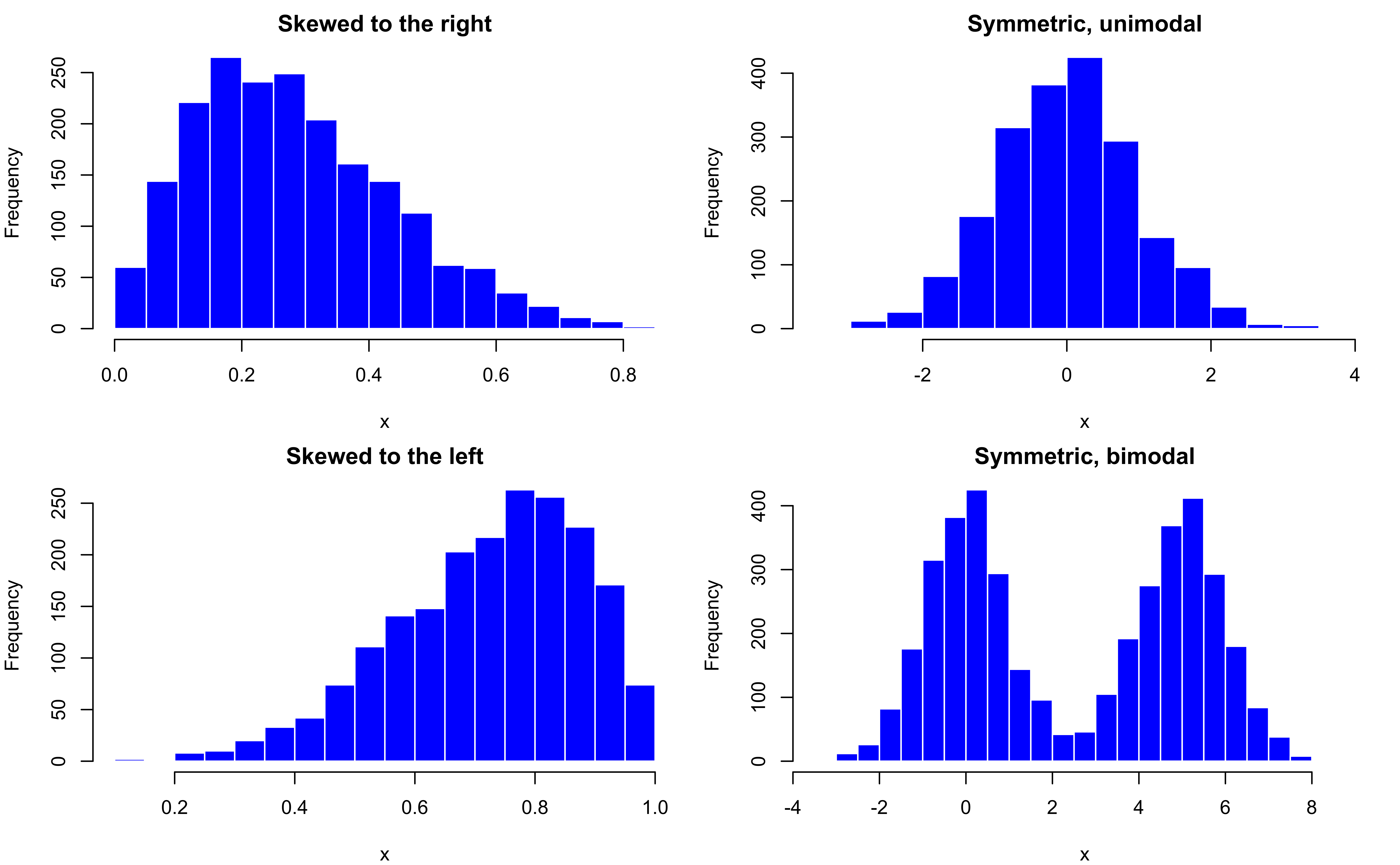
Remembering Skewness
Is the body mass histogram left skewed or right skewed?
Scatterplot for Two Numerical Variables
- A scatterplot provides a case-by-case view of data for two numerical variables.
plot(x = penguins$bill_length_mm, y = penguins$bill_depth_mm,
xlab = "Bill Length", ylab = "Bill Depth",
pch = 16, col = 4)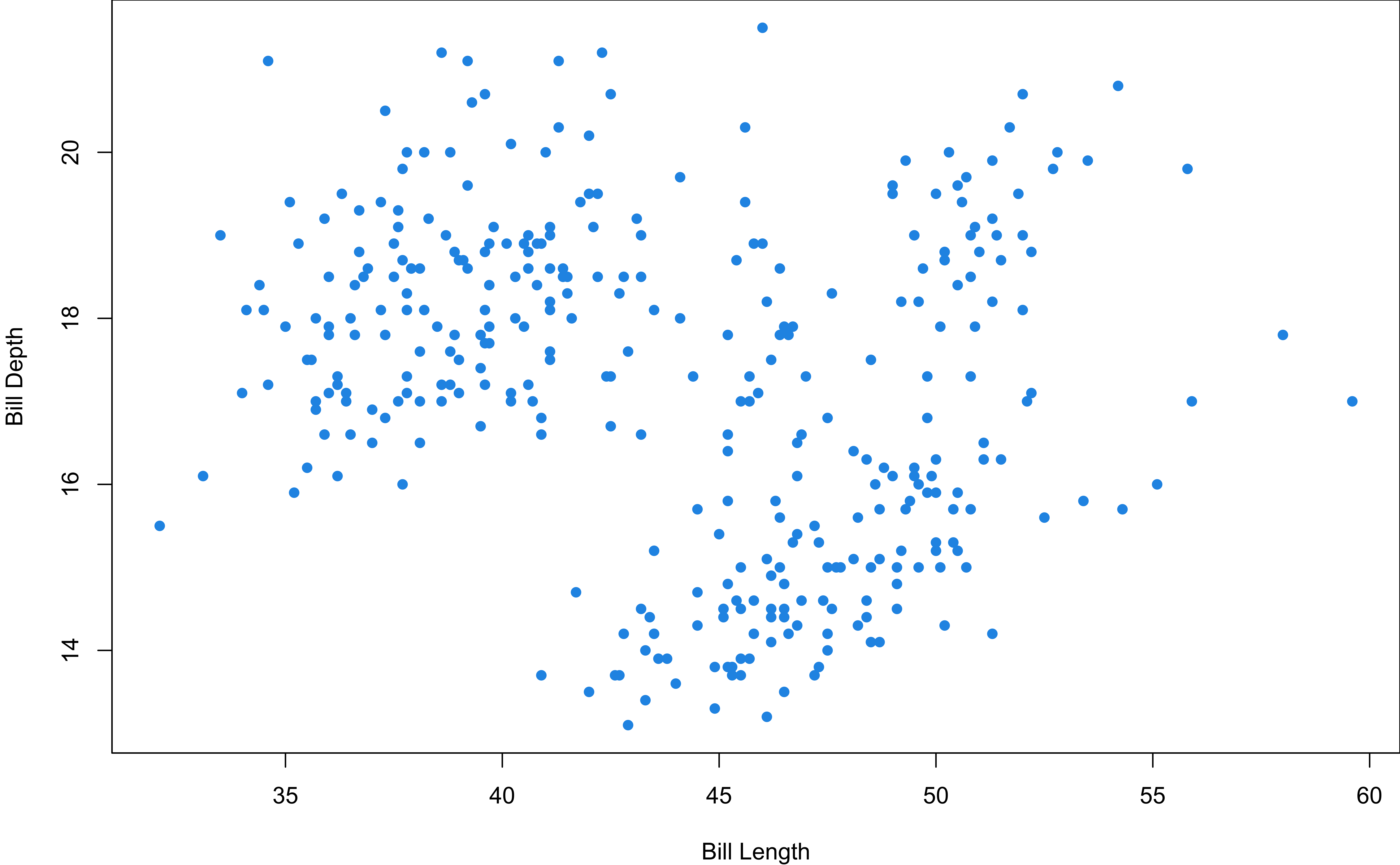
For the penguins data, do
- Pie chart for variable
island. - Histogram for variable
flipper_length_mm. Discuss its shape. - Scatter plot for variables
flipper_length_mmandbill_length_mm.
02:00
Numerical Summaries of Data

If you need to choose one value that represents the entire data, what value would you choose?
Measure of Center: We typically use the middle point. (What does “middle” mean?)
Measure of Variation: What values tell us how much variation a variable has?
Measures of Center: Mean
The (arithmetic) mean or average is adding up all of the values, then dividing by the total number of them.
Let \(x_1, x_2, \dots, x_n\) denote the measurements observed in a sample of size \(n\). Then the sample mean is defined as \[\overline{x} = \frac{\sum_{i=1}^{n} x_i}{n} = \frac{x_1 + x_2 + \dots + x_n}{n}\]
In the body mass example, \[\overline{x} = \frac{3750 + 3800 + \cdots + 3775}{342} = 4202\]
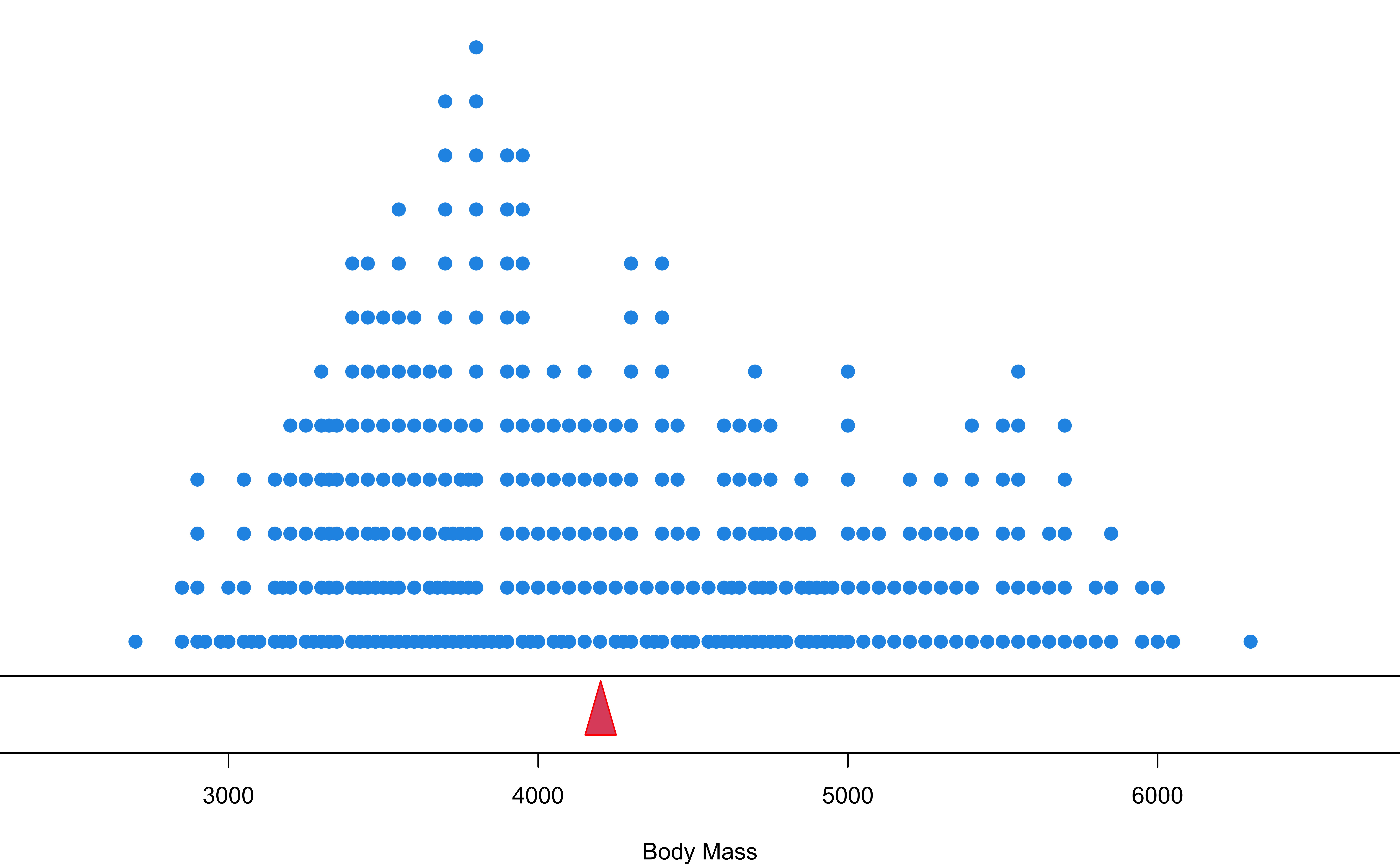
mean(body_mass)[1] 4202Balancing Point
- Think of the mean as the balancing point of the distribution.

Measures of Center: Median
Median: the middle value when data values are sorted.
Half of the values are less than or equal to the median, and the other half are greater than it.
To find the median, we first sort the values.
-
\(n\) is odd: the median is located in the exact middle of the ordered values.
- Data: (0, 2, 10, 14, 8)
- Sorted Data: (0, 2, 8, 10, 14)
- The median is \(8\)
-
\(n\) is even: the median is the average of the two middle numbers.
- Data: (0, 2, 10, 14, 8, 12)
- Sorted Data: (0, 2, 8, 10, 12, 14)
- The median is \(\frac{8 + 10}{2} = 9\)
Calculate Median in R
median(body_mass) ## Compute the median using command median()[1] 4050(sort_mass <- sort(body_mass)) ## sort data [1] 2700 2850 2850 2900 2900 2900 2900 2925 2975 3000 3000 3050 3050 3050 3050
[16] 3075 3100 3150 3150 3150 3150 3175 3175 3200 3200 3200 3200 3200 3250 3250
[31] 3250 3250 3250 3275 3300 3300 3300 3300 3300 3300 3325 3325 3325 3325 3325
[46] 3350 3350 3350 3350 3350 3400 3400 3400 3400 3400 3400 3400 3400 3425 3425
[61] 3450 3450 3450 3450 3450 3450 3450 3450 3475 3475 3475 3500 3500 3500 3500
[76] 3500 3500 3500 3525 3525 3550 3550 3550 3550 3550 3550 3550 3550 3550 3575
[91] 3600 3600 3600 3600 3600 3600 3600 3625 3650 3650 3650 3650 3650 3650 3675
[106] 3675 3700 3700 3700 3700 3700 3700 3700 3700 3700 3700 3700 3725 3725 3725
[121] 3750 3750 3750 3750 3750 3775 3775 3775 3775 3800 3800 3800 3800 3800 3800
[136] 3800 3800 3800 3800 3800 3800 3825 3850 3875 3900 3900 3900 3900 3900 3900
[151] 3900 3900 3900 3900 3950 3950 3950 3950 3950 3950 3950 3950 3950 3950 3975
[166] 4000 4000 4000 4000 4000 4050 4050 4050 4050 4050 4050 4075 4100 4100 4100
[181] 4100 4100 4150 4150 4150 4150 4150 4150 4200 4200 4200 4200 4200 4250 4250
[196] 4250 4250 4250 4275 4300 4300 4300 4300 4300 4300 4300 4300 4350 4350 4375
[211] 4400 4400 4400 4400 4400 4400 4400 4400 4450 4450 4450 4450 4450 4475 4500
[226] 4500 4500 4550 4550 4575 4600 4600 4600 4600 4600 4625 4625 4650 4650 4650
[241] 4650 4650 4675 4700 4700 4700 4700 4700 4700 4725 4725 4725 4750 4750 4750
[256] 4750 4750 4775 4800 4800 4800 4850 4850 4850 4850 4875 4875 4875 4900 4900
[271] 4925 4925 4950 4950 4975 5000 5000 5000 5000 5000 5000 5050 5050 5050 5100
[286] 5100 5100 5150 5150 5200 5200 5200 5200 5250 5250 5250 5300 5300 5300 5300
[301] 5350 5350 5350 5400 5400 5400 5400 5400 5450 5500 5500 5500 5500 5500 5550
[316] 5550 5550 5550 5550 5550 5600 5600 5650 5650 5650 5700 5700 5700 5700 5700
[331] 5750 5800 5800 5850 5850 5850 5950 5950 6000 6000 6050 6300length(body_mass) ## Check sample size is odd or even[1] 342(sort_mass[171] + sort_mass[172]) / 2 ## Verify the answer[1] 4050(body_mass[171] + body_mass[172]) / 2 ## Using un-sorted data leads to a wrong answer!![1] 5525Measures of Center: Mode
Mode: the value that occurs most frequently.
For continuous numerical data, it is common to have no observations with the same value.
Practical definition: A mode is represented by a prominent peak in the distribution.
## Create a frequency table
table_data <- table(body_mass)## Sort the table to find the mode that occurs most frequently
## the number that happens most frequently will be the first one
sort(table_data, decreasing = TRUE)body_mass
3800 3700 3900 3950 3550 3400 3450 4300 4400 3500 3600 3300 3650 4050 4150 4700
12 11 10 10 9 8 8 8 8 7 7 6 6 6 6 6
5000 5550 3200 3250 3325 3350 3750 4000 4100 4200 4250 4450 4600 4650 4750 5400
6 6 5 5 5 5 5 5 5 5 5 5 5 5 5 5
5500 5700 2900 3050 3150 3775 4850 5200 5300 3475 3725 4500 4725 4800 4875 5050
5 5 4 4 4 4 4 4 4 3 3 3 3 3 3 3
5100 5250 5350 5650 5850 2850 3000 3175 3425 3525 3675 4350 4550 4625 4900 4925
3 3 3 3 3 2 2 2 2 2 2 2 2 2 2 2
4950 5150 5600 5800 5950 6000 2700 2925 2975 3075 3100 3275 3575 3625 3825 3850
2 2 2 2 2 2 1 1 1 1 1 1 1 1 1 1
3875 3975 4075 4275 4375 4475 4575 4675 4775 4975 5450 5750 6050 6300
1 1 1 1 1 1 1 1 1 1 1 1 1 1 Comparison of Mean, Median and Mode
Mean is sensitive to extreme values (outliers).
Median/mode is more robust than mean.
head(data_extreme, 10) [1] 37500 3800 3250 3450 3650 3625 4675 3475 4250 3300Comparison of Mean, Median and Mode
Mode is applicable for both categorical and numerical data, while median and mean work for numerical data only.
There may be more than one mode, but there is only one median and one mean.
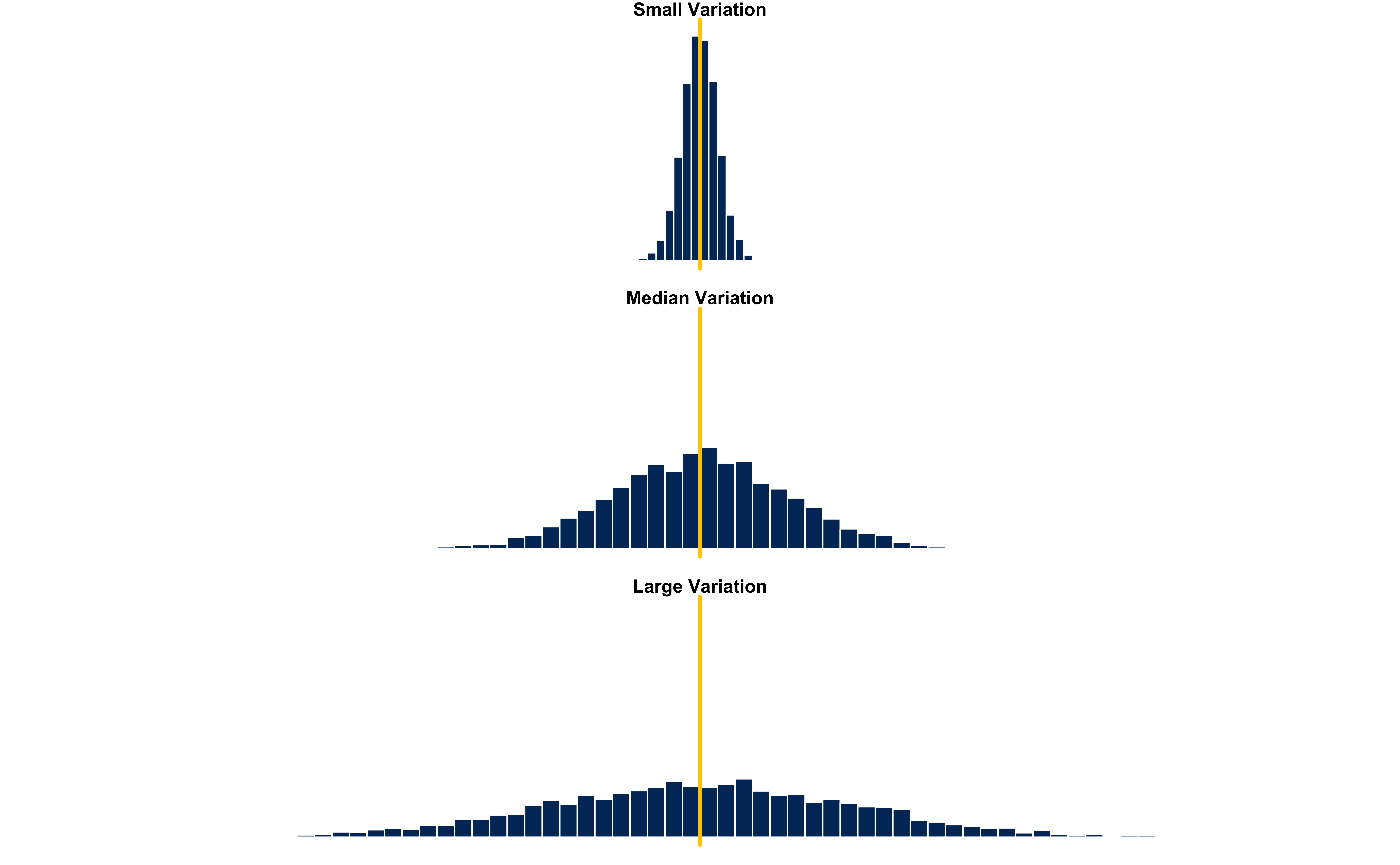
Measures of Variation
Measures of Variation: p-th percentile
-
p-th percentile (quantile): a data value such that
- at most \(p\%\) of the values are below it
- at most \((1-p)\%\) of the values are above it
- Two datasets with the same mean 20.
- One data set has 99-th percentile = 30, and 1-st percentile = 10.
- The other has 99-th percentile = 40, and 1-st percentile = 0.
- Which data have larger variation?
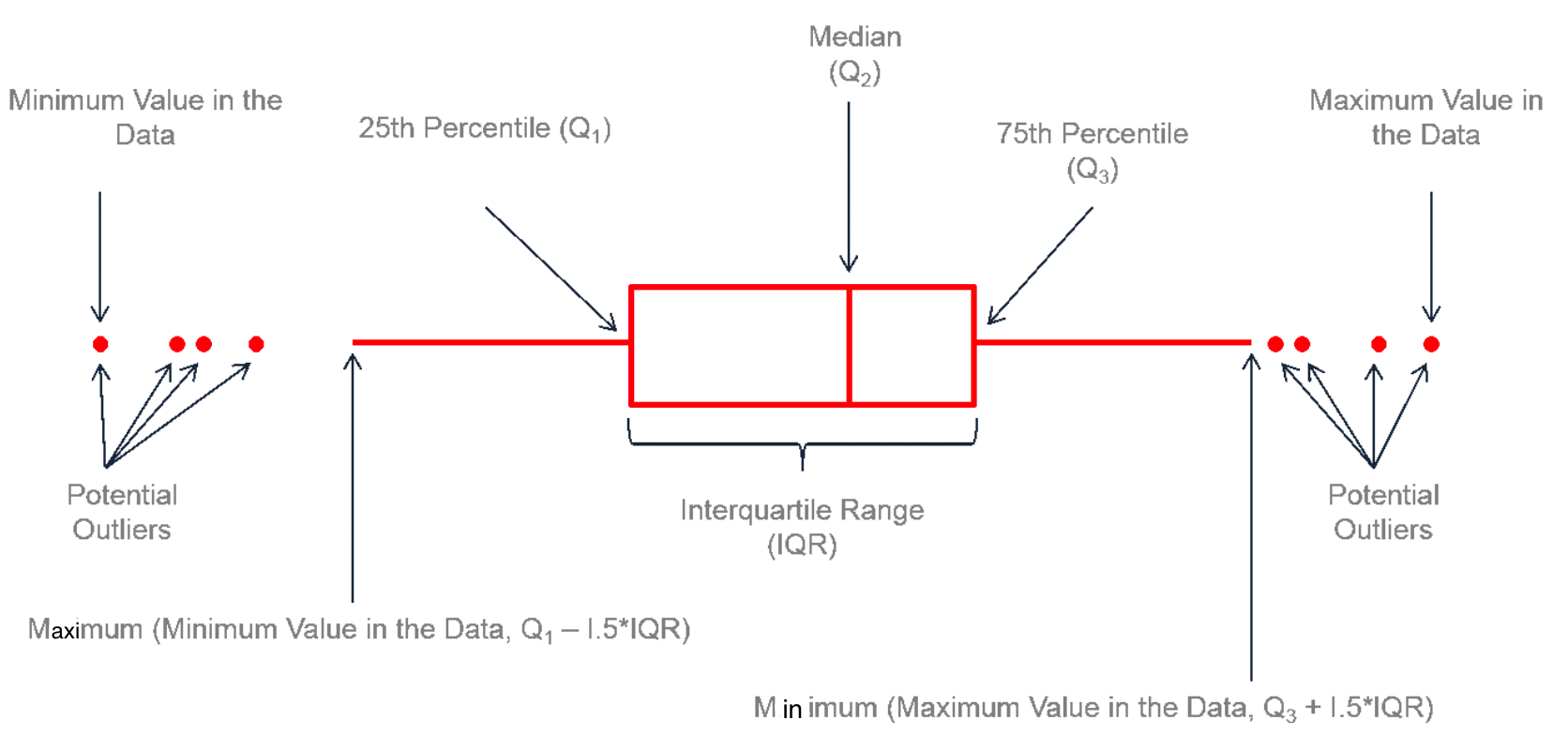
Measures of Variation: Interquartile Range (IQR)
First Quartile (Q1): the 25-th percentile
Second Quartile (Q2): the 50-th percentile (Median)
Third Quartile (Q3): the 75-th percentile
Interquartile Range (IQR): Q3 - Q1
Variance and Standard Deviation
The distance of an observation from its mean, \(x_i - \overline{x}\), its deviation.
Sample Variance is defined as \[ s^2 = \frac{\sum_{i=1}^n(x_i - \overline{x})^2}{n-1} \]
Sample Standard Deviation (SD) is defined as the square root of the variance \[ s = \sqrt{\frac{\sum_{i=1}^n(x_i - \overline{x})^2}{n-1}} \]
Variance is the average of squared deviation from the sample mean \(\overline{x}\) or the mean squared deviation from the mean.
SD is the root mean squared deviation from the mean. It measures, on average, how far the data spread out around the average.
Compute Variance and SD
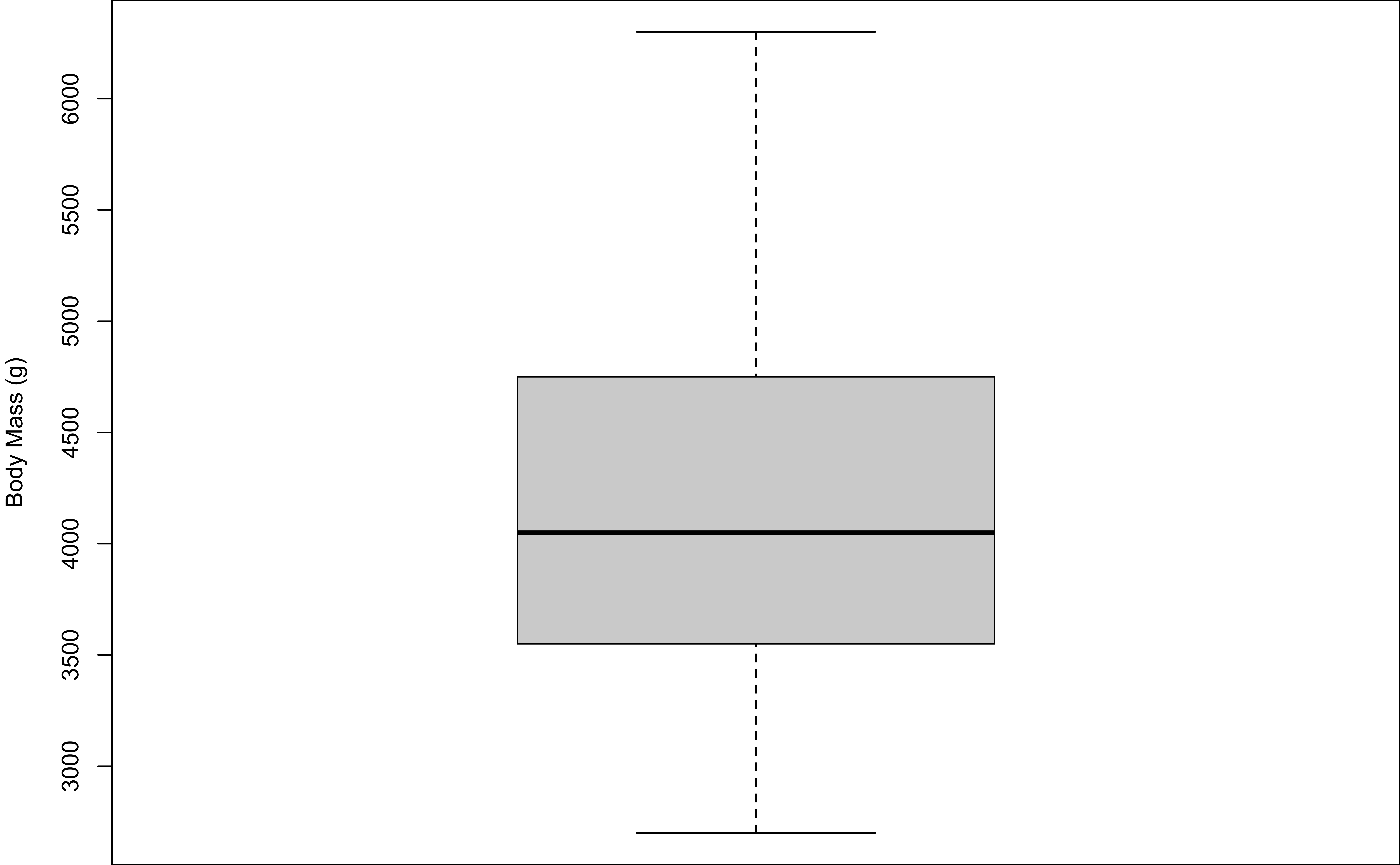
Visualizing Data Variation: Boxplot
When plotting the whiskers,
minimum value in the data means the minimal value that is not an potential outlier.
maximum value in the data means the maximal value that is not an potential outlier.
Body Mass Boxplot
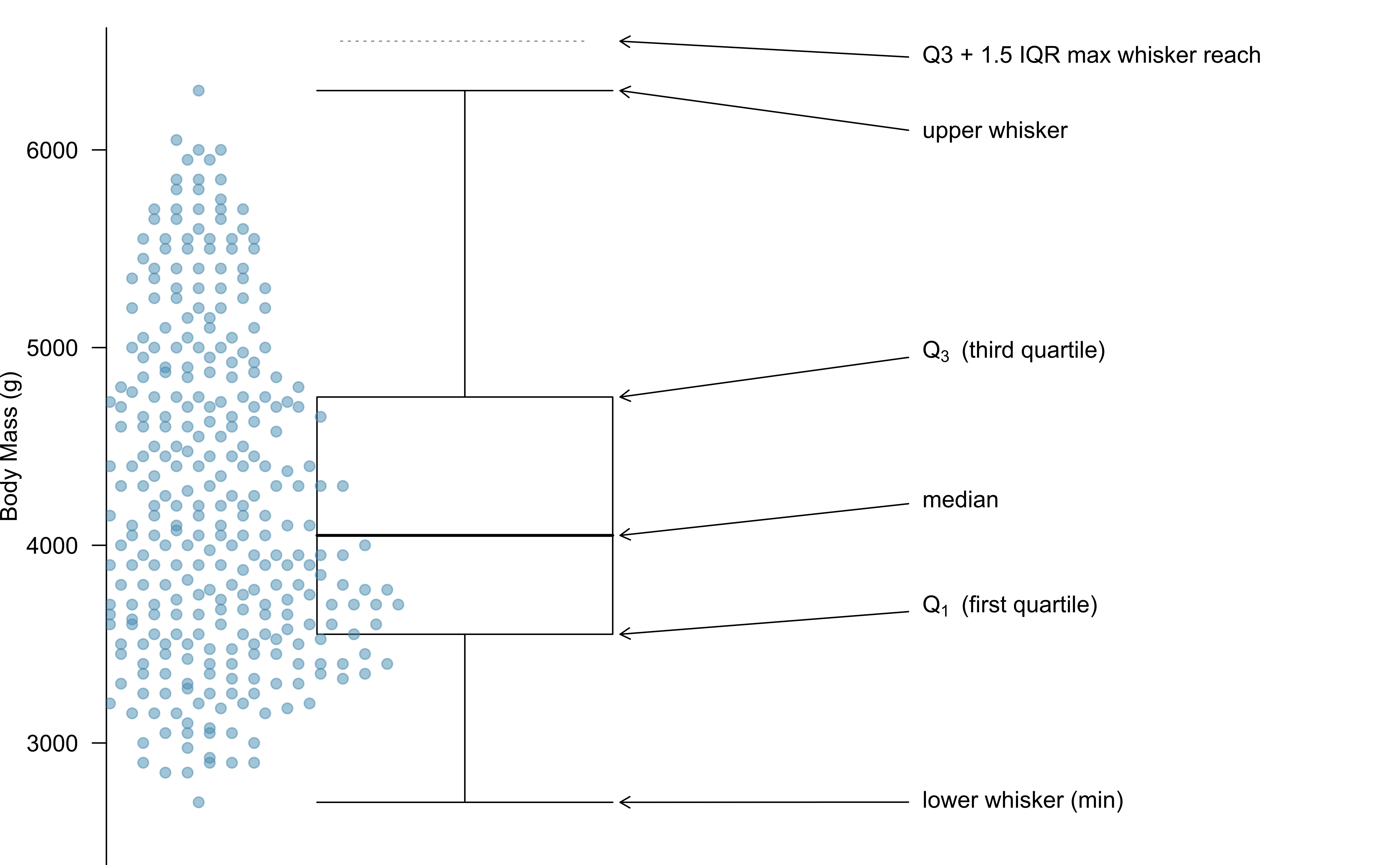
Boxplot in R
For the penguins data,
Make a boxplot for the variable
bill_depth_mm.Compute the minimum, Q1, Q2, Q3, and maximum values of
bill_depth_mm. (Hint:summary()function is pretty useful! 👍)
02:00